Bisulfite Conversion and Library Prep of MBD Enriched Samples (with TapeStation Results)
Library Prep using Zymo Pico Methyl Seq Kit of MBD Enriched FACE_PUF Samples from Previous Post I, Previous Post II, and Previous Post III
First step is to calculate how much DNA in each sample we have so that we can calculate the amount of non-methylated eColi DNA to spike into each sample to check for bisulfite conversion efficiency. We have that DNA diluted to 2.5ng/µl and we should spike 5ng for ever 100ng of sample. Should have done this but I was not aware of it beforehand
Before starting, needed to calculate how the volume of DNA input from the Capture Fraction of MBD Enriched DNA. From the DNA Input Tests found that 1ng of DNA was the optimal amount of DNA to start this library prep kit with.
Example: ACR_790 = 0.545ng/uL ; (1ng)/(0.545ng/uL) = 1.83uL
| Sample | DNA Conc (ng/uL) | Volume of Input DNA (uL) | Volume of Water (uL) |
|---|---|---|---|
| ACR_790 | 0.545 | 1.83 | 18.17 |
| ACR_185 | 1.55 | 0.65 | 19.35 |
| ACR_173 | 0.246 | 4.07 | 15.93 |
| ACR_754 | 0.387 | 2.58 | 17.42 |
| ACR_161 | 0.523 | 1.91 | 18.09 |
| ACR_13 | 0.303 | 3.30 | 16.70 |
| ACR_778 | 0.562 | 1.78 | 18.22 |
| ACR_209 | 0.197 | 5.08 | 14.92 |
| ACR_221 | 0.508 | 1.97 | 18.03 |
Section 1: BS Conversion Conversion Reaction
- 20µl of each tube was taken and pipetted into new PCR tubes, one for each sample
- 130µl of Lightning Conversion Reagent was added to each tube, vortexed, then spun down
- The 5 tubes were put into the thermocycler bisulfite conversion program (under mes user), which is 98 degrees C for 8 minutes, then 54 degrees C for 60 minutes, then 4 degree C hold
Conversion Cleanup
- Added 600µl of M-Binding buffer (lightning kit) to the provided Zymo-Spin IC Columns (4 of them, one per sample)
- Added the bisulfite treated DNA (150µl) to its respective column and inverted to mix
- Centrifuged at 12,000 rcf for 30 seconds and discarded flowthrough
- Added 100µl M-wash buffer to each column and centrifuged at 12,000 rcf for 30 seconds
- Discarded flowthrough
- Added 200µl L-Desulfonation buffer to each column and let them sit at room temp for 15 minutes
- Began warming elution buffer to 56 degrees C
- Centrifuged for 30 seconds at 12,000 rcf and discarded flowthrough
- Added 200µl M-wash buffer, centrifuged for 30 seconds at 12,000 rcf and discarded flowthrough
- Repeated above wash step one time
- Transferred columns to 1.5mL tubes
- Added 8µl warmed elution buffer to the columns and let them sit for 1 minute
- Centrifuged for 30 seconds at 12,000 rcf
Section 2: Amplification with PrepAMP Primers
- Made Priming master mix on ice:
- 2µl 5X PrepAmp buffer * 9.2 = 18.4µl
- 1µl PrepAmp Primers (40µM) * 9.2 = 9.2µl
- Made new PCR tubes with 3µl of PrepAmp MM and 7µl of bisulfite treated DNA (100ng, 10ng, 1ng, 100pg, 10pg)
- Kept those on ice
- Made PrepAmp Mix on ice:
- 1µl 5X PrepAmp buffer * 9.2 = 9.2µl
- 3.75µl PrepAmp PreMix * 9.2 = 34.5uL
- 0.3µl PrepAmp polymerase * 9.2 = 2.76µl
- Set thermocylcer program with lid temp restricted to 25 degrees C and place samples inside and run:
- 98 for 2 minutes
- 8 degrees for 1 minute
- 8 degree hold
- During hold vortex, spin tubes down, add 5.05µl PrepAmp Mix to each tube, vortex, spin down, and place back in thermocycler
- 8 degrees for 4 minutes
- 16 degrees for 1 minute with 3% ramp rate
- 22 degrees for 1 minute with 3% ramp rate
- 28 degrees for 1 minute with 3% ramp rate
- 36 degrees for 1 minute with 3% ramp rate
- 36.5 degrees for 1 minute with 3% ramp rate
- 37 degrees for 8 minutes
- repeat back from the first step one time through again
- During hold, vortex, spin tubes down, add 0.3µl PrepAmp Polymerase to each tube, vortex, spin down, and place back into thermocycler
Section 3: DNA Clean and Concentrator Columns (DCC)
- Made a 1.5mL tube for each sample, added 7:1 ratio DNA binding buffer, so 250.81µl (Should have been 107.45uL) of DNA binding buffer and 15.35uL of Product
- Put elution buffer in thermomixer 56 degrees
- Added DNA sample (15.35µl) to the appropriate 1.5mL tube
- Vortexed, spun down, and added to the column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Transferred columns to 1.5mL tubes
- Added 12µl warmed elution buffer to each column directly
- Incubated 1 minute
- Centrifuged 12,000 rcf 30 seconds
Section 4: Second Amplification (Adapater Addition)
- Made 1st Amp master mix:
- 12.5µl 2X Library Amp Mix * 9.2 = 115µl
- 1µl Library Amp Primer(10µM) * 9.2 = 9.2µl
- Added 13.5µl MM to new PCR tubes
- Added 11.5µl of cleaned and concentrated DNA sample to the appropriate new PCR tube
- Vortexed, spun down, and placed in thermocycler programs specific to how many cycles each sample required: -1ng Sample 8 cycles (All these samples were the same ; 1ng of input)
Cleanup with DCC
- Made a 1.5mL tube for each sample, added 7:1 ratio DNA binding buffer, so 175µl of DNA binding buffer
- Put elution buffer in thermomixer 56 degrees
- Added DNA sample (25µl) to the appropriate 1.5mL tube
- Vortexed, spun down, and added to the column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Transferred columns to 1.5mL tubes
- Added 12.5µl warmed elution buffer to each column directly
- Incubated 1 minute
- Centrifuged 12,000 rcf 30 seconds
Section 5: Amplification with Index Primers
- Made new PCR tubes for each sample with 12.5µl Library Amp Mix
- Added indexed primers: -ACR_790 Sample got 0.5uL of Index A -ACR_185 Sample got 0.5uL of Index B -ACR_173 Sample got 0.5uL of Index C -ACR_754 Sample got 0.5uL of Index D -ACR_161 Sample got 0.5uL of Index E -ACR_13 Sample got 0.5uL of Index F -ACR_778 Sample got 0.5uL of Index G -ACR_209 Sample got 0.5uL of Index H -ACR_221 Sample got 0.5uL of Index I Samples that got Indexes G, H and I used Index Primers from RRBS Kit, all rest were used from the Pico Kit
- Added 12µl of sample to the appropriate tube (all of the flowthrough from above DCC)
- Vortexed, spun down, and placed in theremocycler program 2nd Pico Methyl Amp program
Cleanup with DCC
- Made a 1.5mL tube for each sample, added 7:1 ratio DNA binding buffer, so 175µl of DNA binding buffer
- Put elution buffer in thermomixer 56 degrees
- Added DNA sample (25µl) to the appropriate 1.5mL tube
- Vortexed, spun down, and added to the column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Transferred columns to 1.5mL tubes
- Added 12µl warmed elution buffer to each column directly
- Incubated 1 minute
- Centrifuged 12,000 rcf 30 seconds
D5000 TapeStation
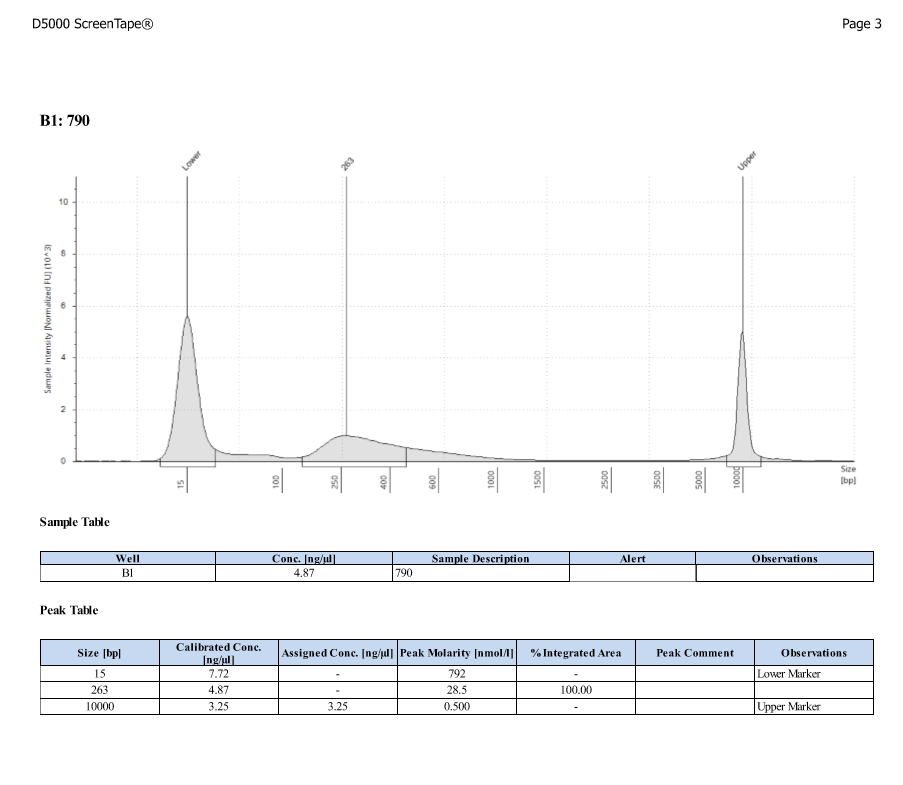
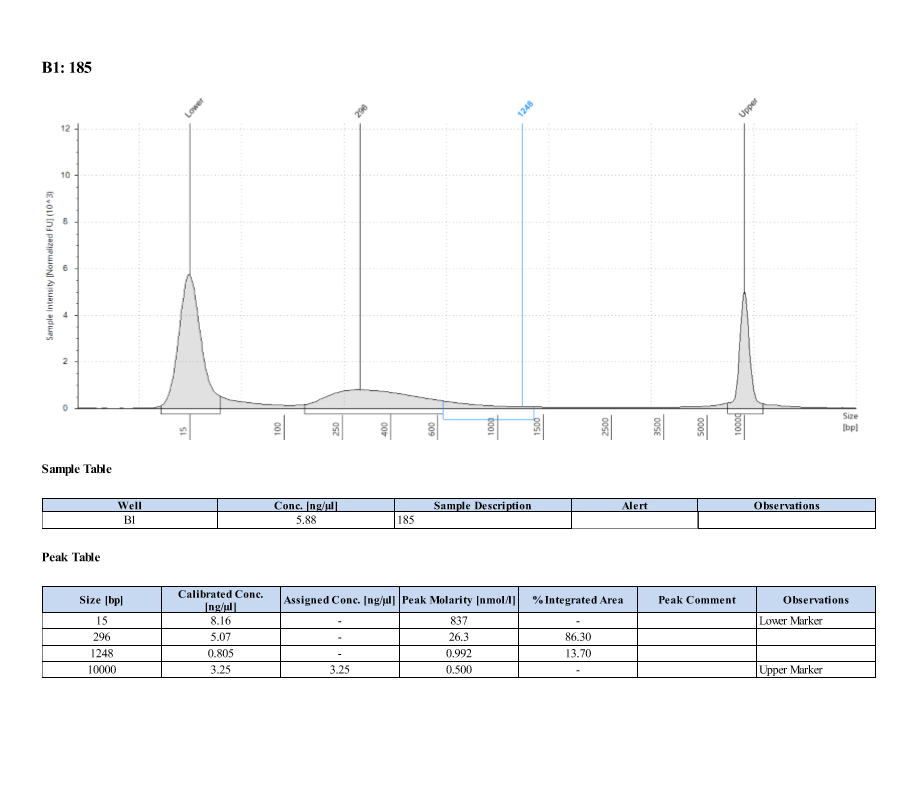
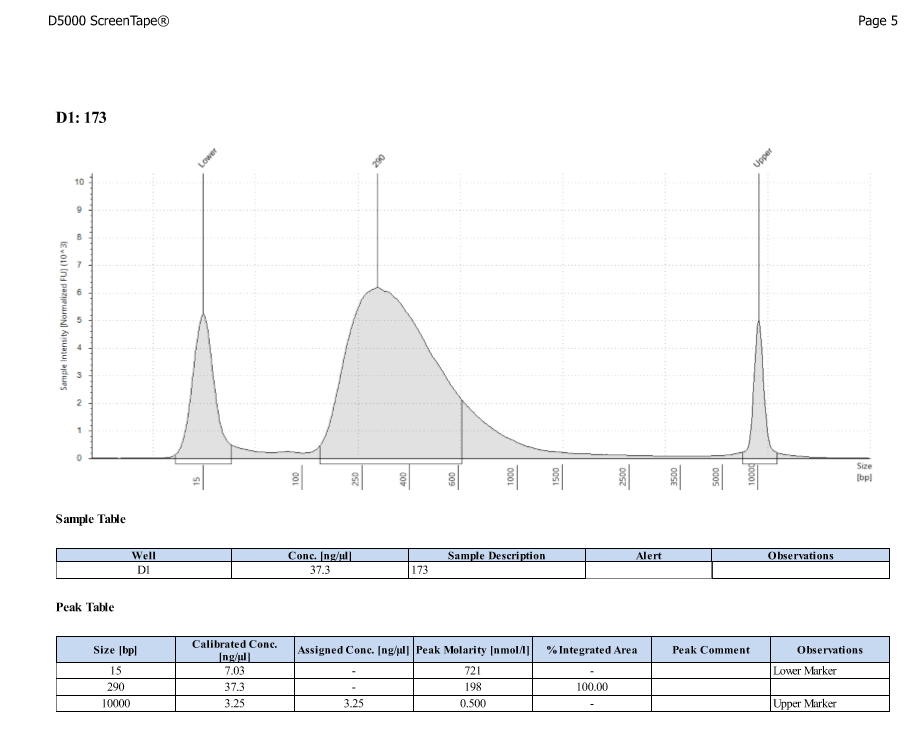
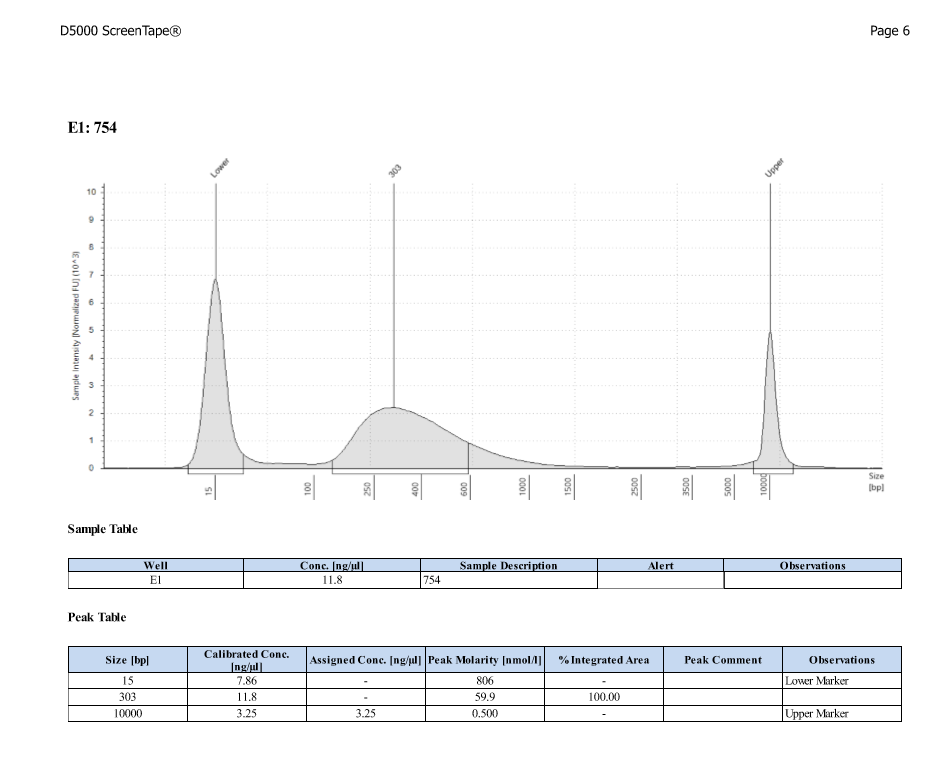
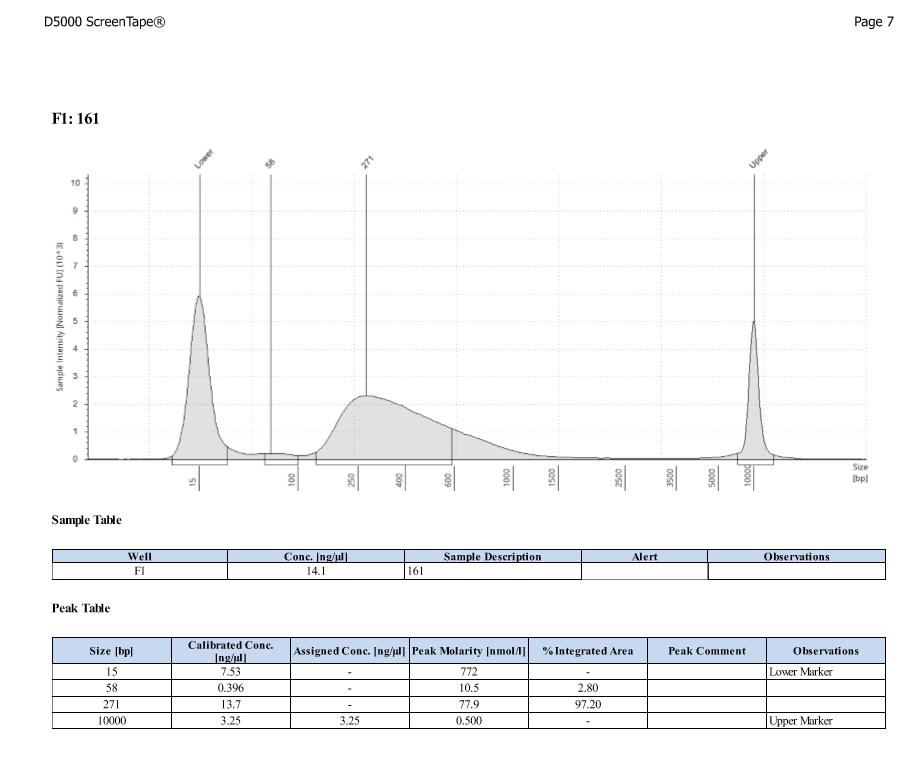
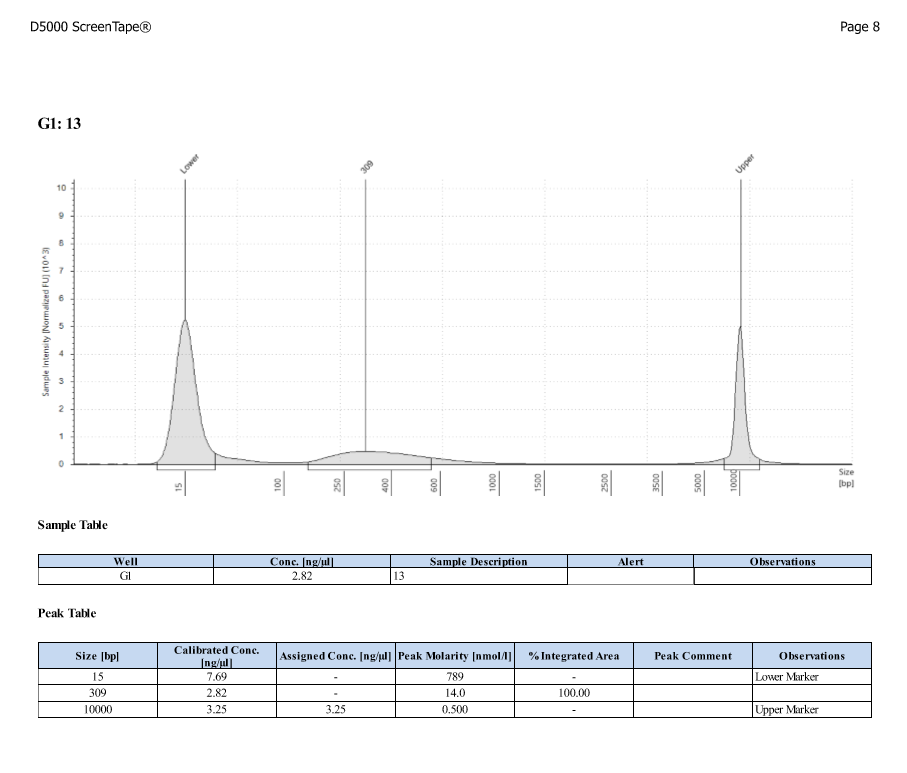
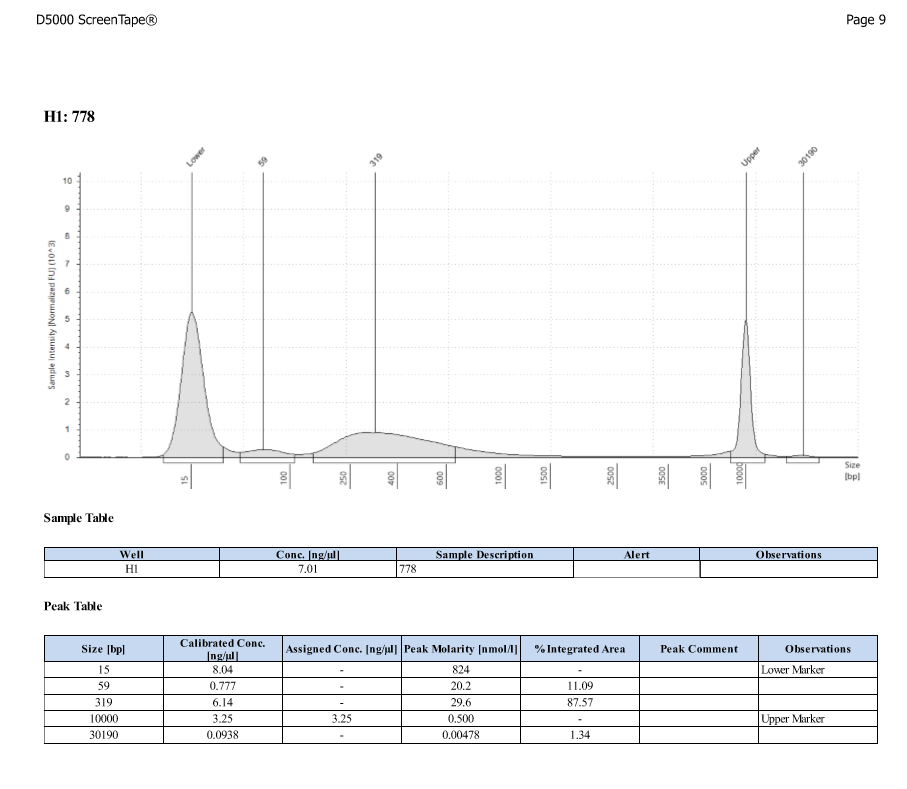
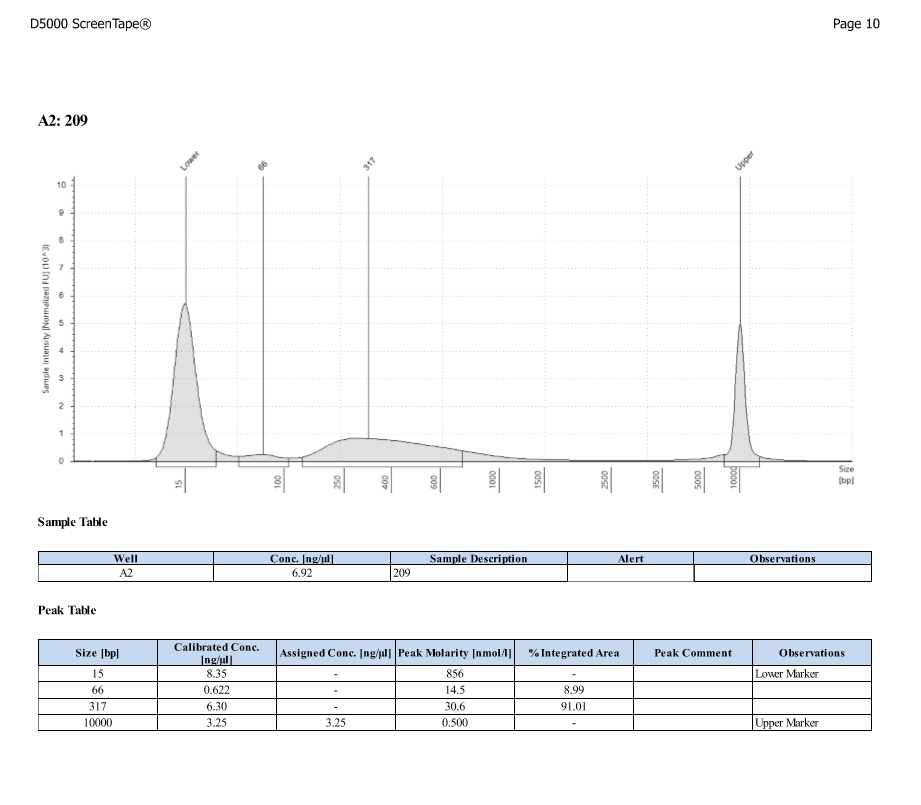
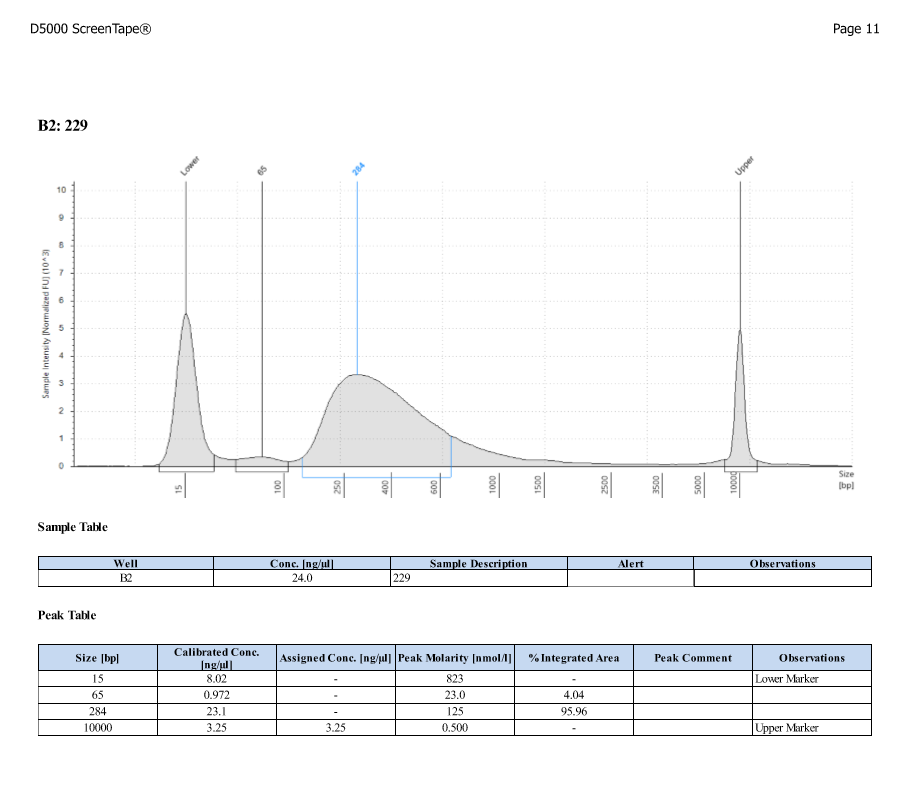
- Protocol for D5000 tapestation followed exactly
- See full results 20190828
- See full results 20190829_790 See full results 20190829_185
