Shearing FACE_PUF DNA for Pico Methyl Seq Library
Shearing FACE_PUF ACR Sperm Samples Using Q800R3 Sonicator via DNA Shearing Protocol
Using the Qubit Values from Erin’s Extractions 1 and Erin’s Extractions 2 I calculated how much volume of sample DNA I needed to input to get 1ug of DNA in 50uL of input volume.
Summary Table of Qubit Results
| Sample Id | Sample Type | Avg Concentration (ng/uL) |
|---|---|---|
| ACR_790 | Sperm | 58.3 |
| ACR_185 | Sperm | 75.9 |
| ACR_173 | Sperm | 87.7 |
| ACR_754 | Sperm | 53.2 |
| ACR_221 | Sperm | 132.0 |
| ACR_161 | Sperm | 45.9 |
| ACR_13 | Sperm | 30.1 |
| ACR_197 | Sperm | 5.35 |
| ACR_778 | Sperm | Too Low |
| ACR_209 | Sperm | 82.0 |
Each input value was calculated by dividing 1000ng or 1ug by the average concentration of each sample to get the volume needed to input to the shearing process. The remainder of the 50uL input was supplemented with Ultra Pure Water.
For _Example
Sample 790 had 58.3 ng/ul so (1000ng)/(58.3ng/uL) = 17.15uL of DNA Input
| Sample Id | DNA Input Vol (uL) | Water Input (uL) |
|---|---|---|
| ACR_790 | 17.15 | 32.85 |
| ACR_185 | 13.18 | 36.82 |
| ACR_173 | 11.40 | 38.60 |
| ACR_754 | 18.80 | 31.20 |
| ACR_221 | 7.58 | 42.42 |
| ACR_161 | 21.79 | 28.21 |
| ACR_13 | 33.22 | 16.78 |
| ACR_197 | NA | NA |
| ACR_778 | NA | NA |
| ACR_209 | 12.20 | 37.80 |
Samples 197 and 778 were not run because of their low concentrations (So low that even if used entire sample it would not equal 1ug which was required to start the shearing process) Sample 221 was also not run because it was already sheared ACR_221_Tube9
All samples were sonicated for 4 cycles of 15s on and 15off for a total of 2 minutes at 25% Amplification.
D5000 TapeStation
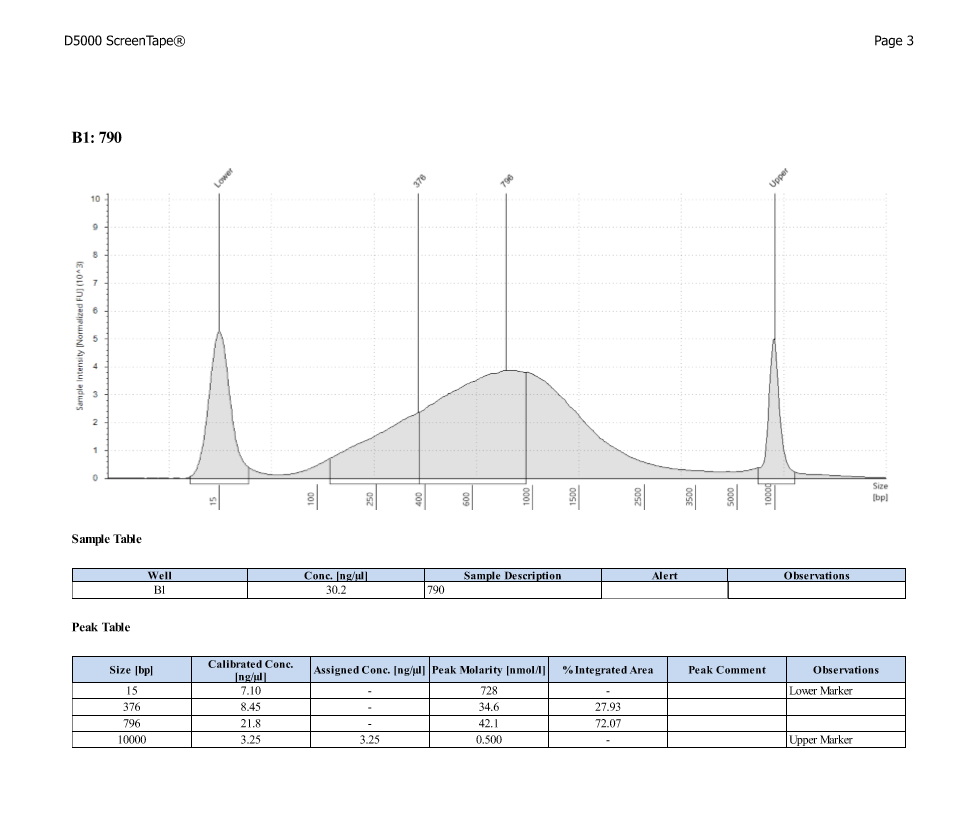
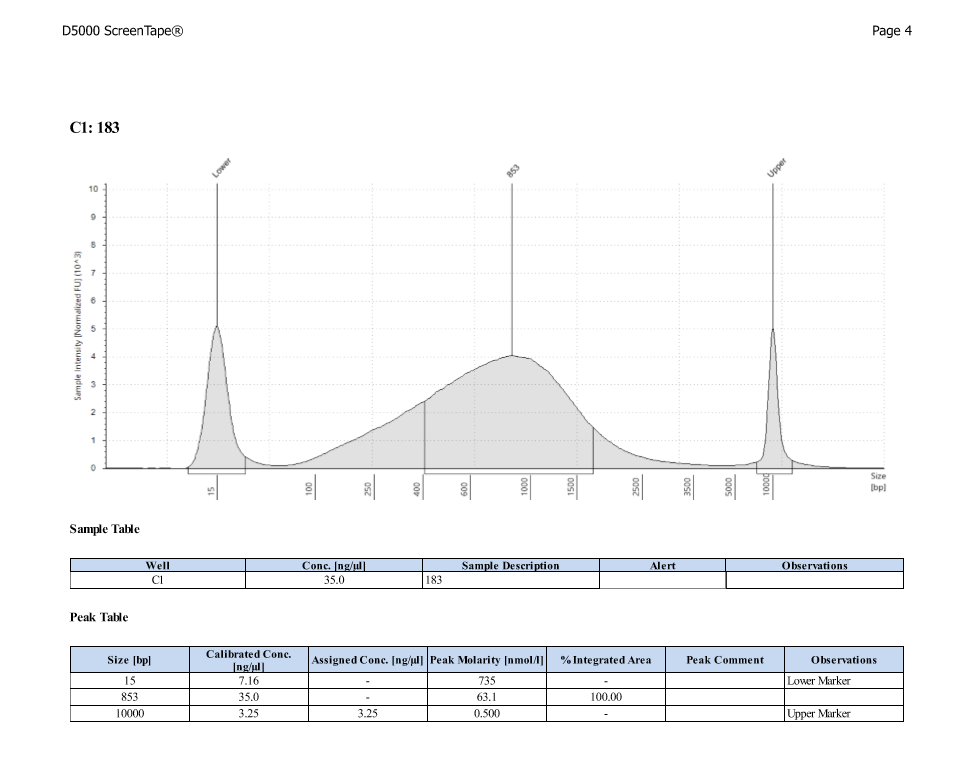
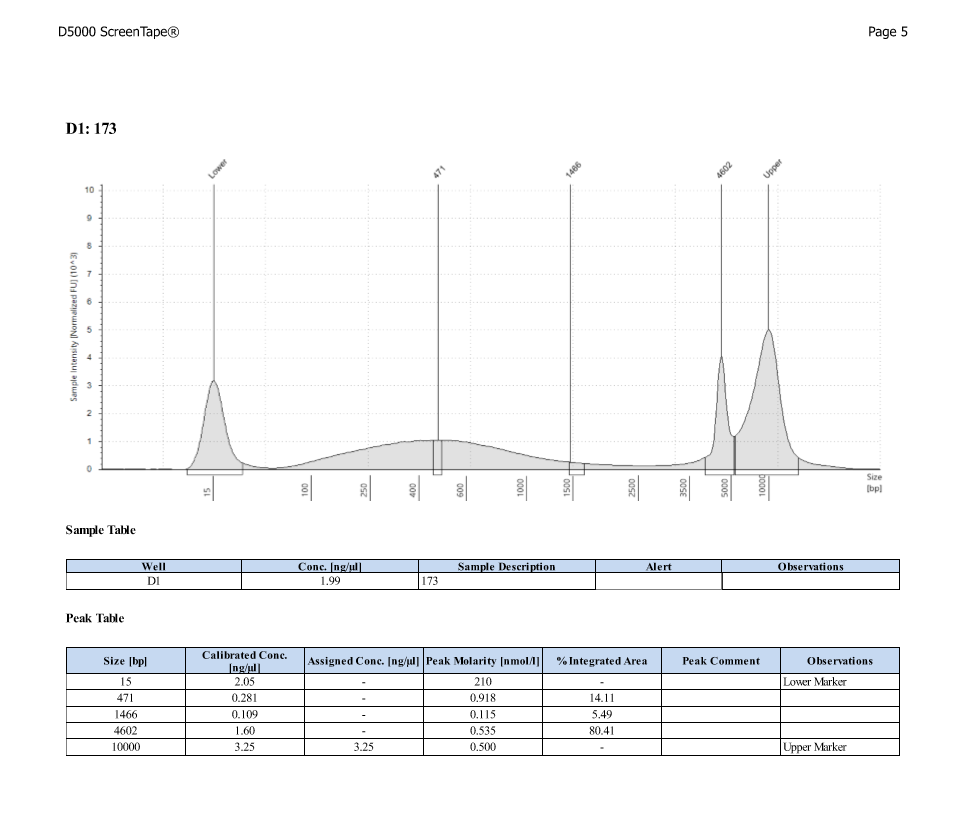
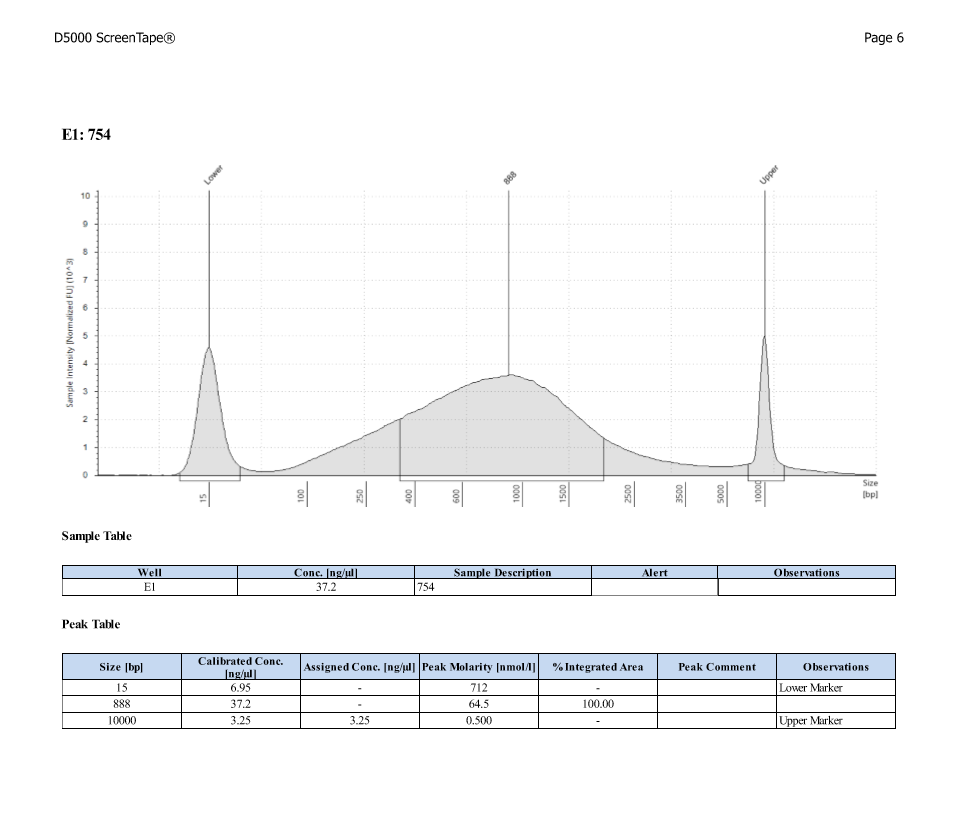
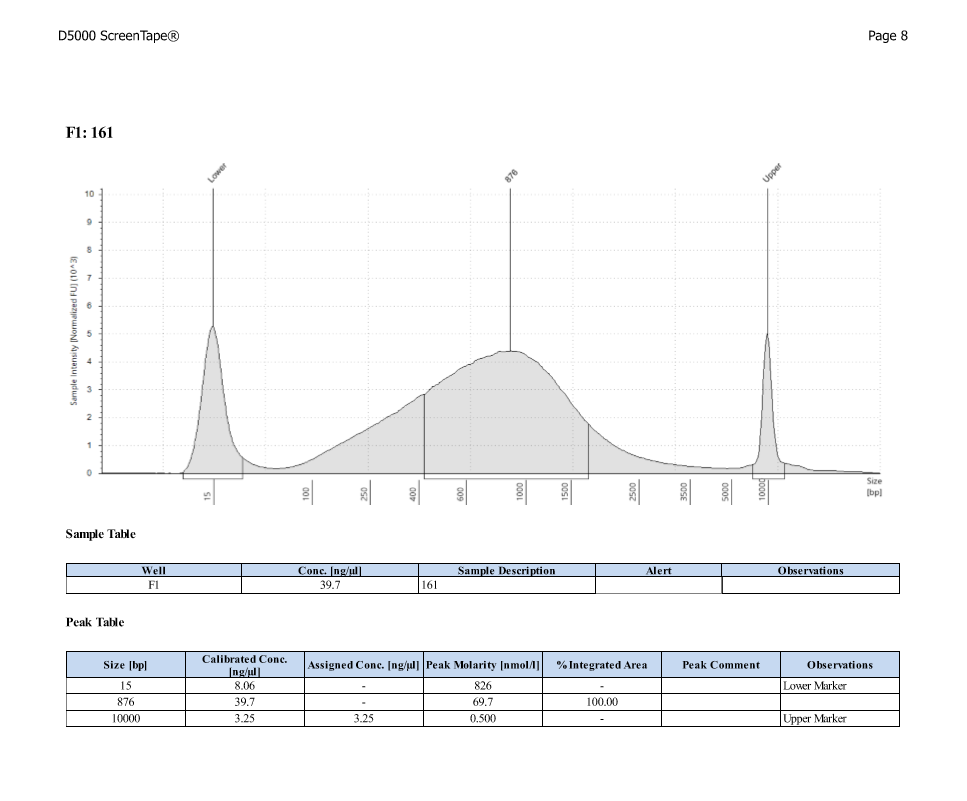
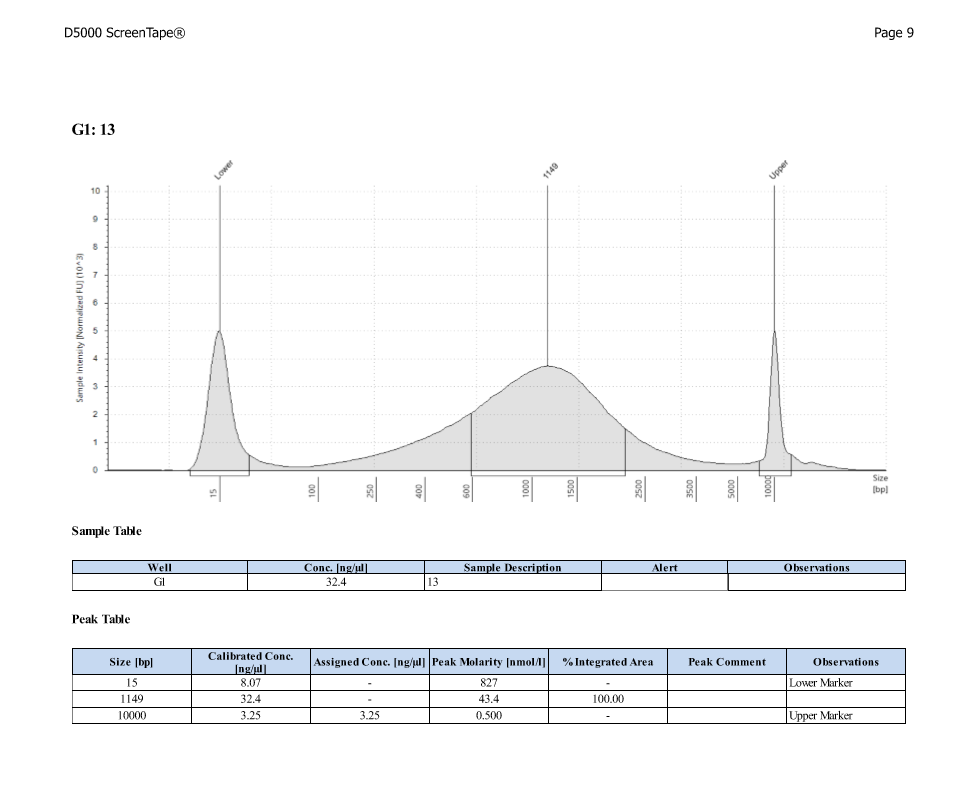
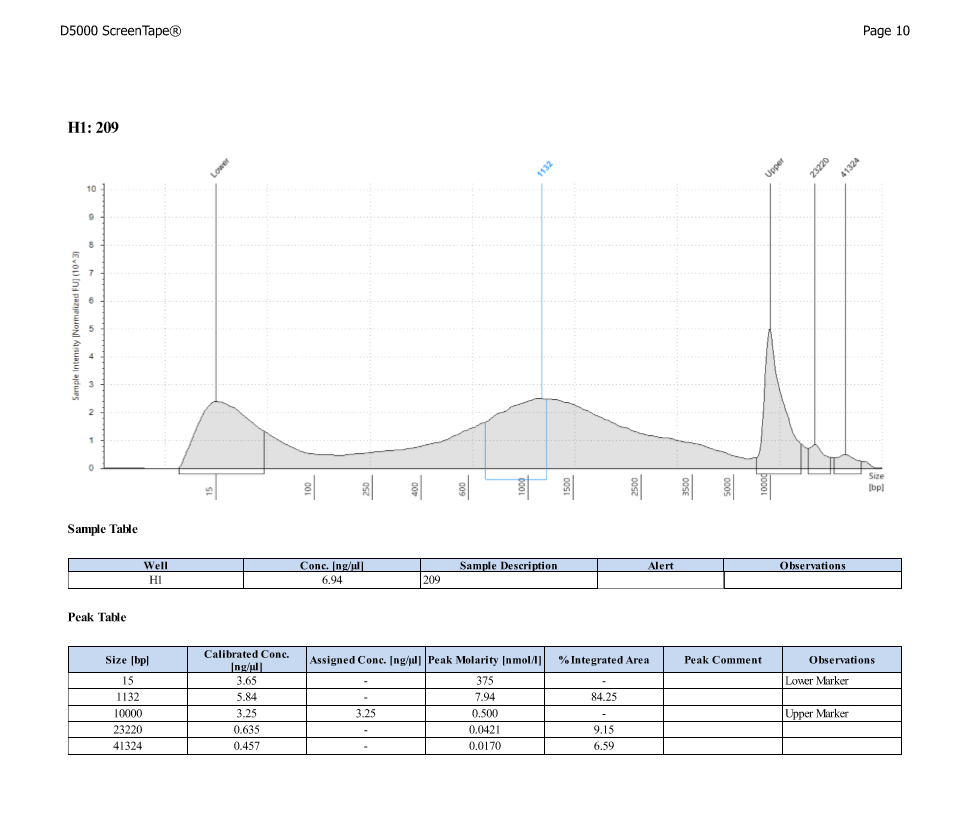
After Sonicating these samples, most of the samples were still out of the desired range of 500-650bp (were all about 800-1100bp range except 173)
